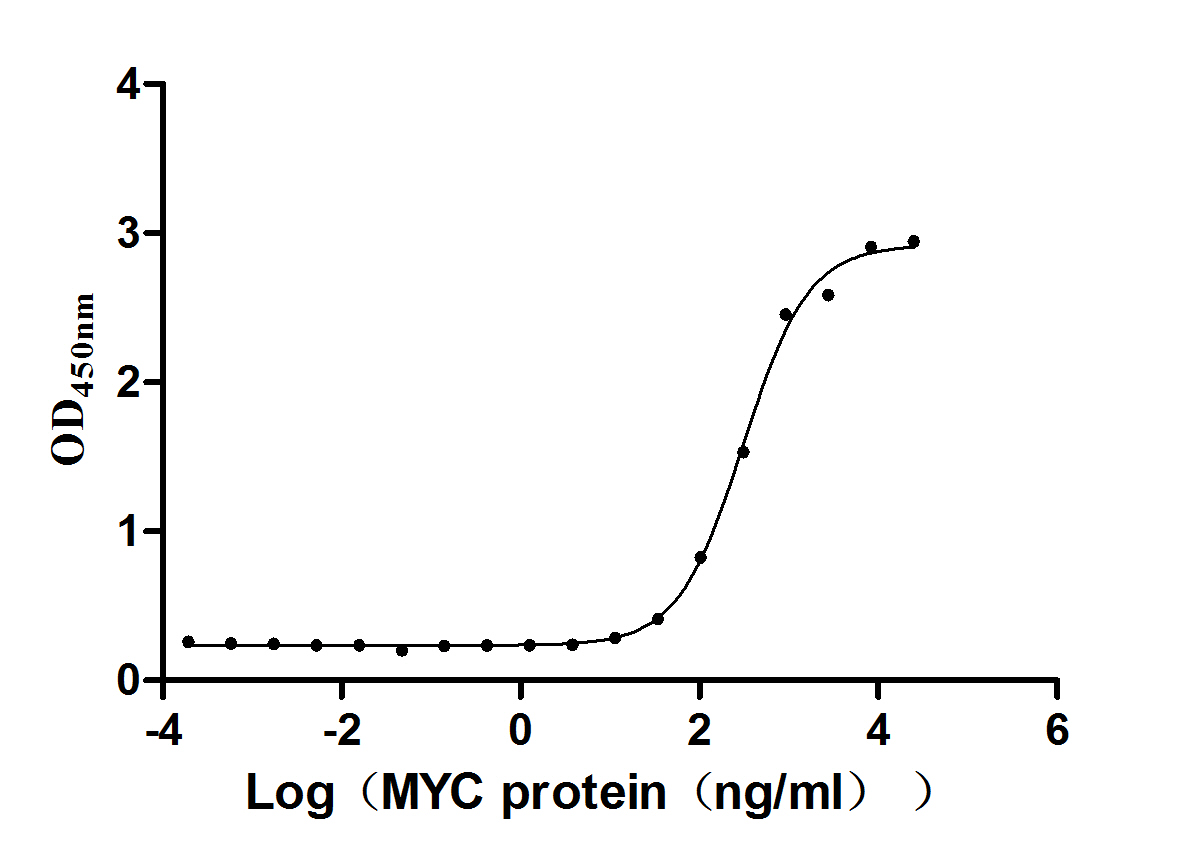
Recombinant Escherichia coli Chaperone protein DnaK (dnaK)
-
中文名稱:大腸桿菌dnaK重組蛋白
-
貨號:CSB-YP363924ENV
-
規(guī)格:
-
來源:Yeast
-
其他:
-
中文名稱:大腸桿菌dnaK重組蛋白
-
貨號:CSB-EP363924ENV
-
規(guī)格:
-
來源:E.coli
-
其他:
-
中文名稱:大腸桿菌dnaK重組蛋白
-
貨號:CSB-EP363924ENV-B
-
規(guī)格:
-
來源:E.coli
-
共軛:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名稱:大腸桿菌dnaK重組蛋白
-
貨號:CSB-BP363924ENV
-
規(guī)格:
-
來源:Baculovirus
-
其他:
-
中文名稱:大腸桿菌dnaK重組蛋白
-
貨號:CSB-MP363924ENV
-
規(guī)格:
-
來源:Mammalian cell
-
其他:
產(chǎn)品詳情
-
純度:>85% (SDS-PAGE)
-
基因名:dnaK
-
Uniprot No.:
-
別名:dnaK; groP; grpF; seg; b0014; JW0013; Chaperone protein DnaK; HSP70; Heat shock 70 kDa protein; Heat shock protein 70
-
種屬:Escherichia coli (strain K12)
-
蛋白長度:Full Length of Mature Protein
-
表達區(qū)域:2-638
-
氨基酸序列GKIIGIDLG TTNSCVAIMD GTTPRVLENA EGDRTTPSII AYTQDGETLV GQPAKRQAVT NPQNTLFAIK RLIGRRFQDE EVQRDVSIMP FKIIAADNGD AWVEVKGQKM APPQISAEVL KKMKKTAEDY LGEPVTEAVI TVPAYFNDAQ RQATKDAGRI AGLEVKRIIN EPTAAALAYG LDKGTGNRTI AVYDLGGGTF DISIIEIDEV DGEKTFEVLA TNGDTHLGGE DFDSRLINYL VEEFKKDQGI DLRNDPLAMQ RLKEAAEKAK IELSSAQQTD VNLPYITADA TGPKHMNIKV TRAKLESLVE DLVNRSIEPL KVALQDAGLS VSDIDDVILV GGQTRMPMVQ KKVAEFFGKE PRKDVNPDEA VAIGAAVQGG VLTGDVKDVL LLDVTPLSLG IETMGGVMTT LIAKNTTIPT KHSQVFSTAE DNQSAVTIHV LQGERKRAAD NKSLGQFNLD GINPAPRGMP QIEVTFDIDA DGILHVSAKD KNSGKEQKIT IKASSGLNED EIQKMVRDAE ANAEADRKFE ELVQTRNQGD HLLHSTRKQV EEAGDKLPAD DKTAIESALT ALETALKGED KAAIEAKMQE LAQVSQKLME IAQQQHAQQQ TAGADASANN AKDDDVVDAE FEEVKDKK
-
蛋白標(biāo)簽:Tag?type?will?be?determined?during?the?manufacturing?process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
產(chǎn)品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
復(fù)溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質(zhì)期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事項:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶點詳情
-
功能:Plays an essential role in the initiation of phage lambda DNA replication, where it acts in an ATP-dependent fashion with the DnaJ protein to release lambda O and P proteins from the preprimosomal complex. DnaK is also involved in chromosomal DNA replication, possibly through an analogous interaction with the DnaA protein. Also participates actively in the response to hyperosmotic shock.
-
基因功能參考文獻:
- While preventing the formation of lethal IpaC aggregates, DnaK promoted the incorporation of IpaC into large and dynamic complexes (LDCs) restricted at the bacterial pole through nucleoid occlusion. Direct and reversible IpaC-DnaK interactions as part of large dynamic complexes (LDCs) drive polarization through nucleoid occlusion. Polar accumulation was associated with DnaKJ-dependent substrate folding. PMID: 29795186
- BAH1, an E3 ligase from plant that has a similar zinc finger domain to DnaJ, can perform block the effect of DnaK on sigma(32) in Escherichia coli. PMID: 29260303
- A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy. PMID: 29669923
- the mutant K-12 DeltadnaK was more sensitive to Ultrasound with c. 2.5 log (CFU per ml) reduction in comparison to their isogenic wild-type E. coli K-12. This indicates that the dnaK gene participates in general stress response and more specifically to hyperosmotic stress. PMID: 28727911
- The results provide new insight into the heterogeneous ensemble of complexes formed by DnaK chaperones. PMID: 28833766
- Taken together, these results suggest that a specific region in the nucleotide-binding domain of DnaK is involved in the interaction with Escherichia coli Hsp90, and this interaction is functionally important. PMID: 28013030
- binding of a peptide substrate to the DnaK-peptide-binding domain leads to prominent lid closure. PMID: 27248857
- the helical lid of DnaK is a highly dynamic unit of the structure in all ligand-bound states. Importantly, we demonstrate that DnaK populates a partially docked state in the presence of ATP and substrate and that this state represents an energy minimum on the DnaK allosteric landscape. PMID: 28428246
- this study shows that DnaK has the potential to modify and enhance immunogenicity when associated with aggregated protein PMID: 27859059
- Fatty acid methyl esters analysis indicated that the amount of unsaturated fatty acid sharply increased and subcellular location prediction analysis showed a marked decrease in transcription of inner-membrane protein genes, which might have triggered the development of aberrant cell shape and susceptibility for some antibiotics in the DeltadnaKJ strain. PMID: 27242140
- we observe an additive effect of DnaK and GroEL chaperones on the evolutionary rates of their common interactors. Finally, we found pronounced similarities in the physicochemical profiles that characterize proteins belonging to DnaK and GroEL interactomes. PMID: 27189986
- For DnaK, acetyl-CoA carboxylase, biotin carboxylase subunit (AccC) and phosphate acetyltransferase (Pta) we also showed a direct role of PpiB in the functional control of these proteins because it increased the measured enzyme activity of each protein and further interfered with DnaK localization and the correct folding of AccC. PMID: 27306110
- Data show that a partially docked DnaK structure is achieved by combining ATP in the the N-terminal nucleotide binding domain (NBD) and peptide in the substrate binding domain (SBD). PMID: 27025773
- This unit describes the procedure for following reactivation of an aggregated enzyme glucose-6-phosphate dehydrogenase mediated by ClpB from Escherichia coli in cooperation with another molecular chaperone, DnaK. PMID: 26836408
- The interaction of DnaK with DnaJ, GrpE, or sigma32 becomes weaker when DnaK is glutathionylated during oxidative stress, and the interaction is restored upon deglutathionylation. PMID: 26823468
- Data suggest that neither DnaK nor GroEL singly can modulate sigma32 stability in vivo; there is ordered network between them, where GroEL acts upstream of DnaK. PMID: 26545493
- analysis of human telomere repeat binding factor 1 (hTRF1) in complex with Escherichia coli Hsp70 (DnaK) PMID: 26240333
- the C-terminal helix of the nucleotide binding domain of the Hsp70 chaperone DnaK is the major determinant of mechanical stability PMID: 26240360
- analysis of DnaJ-DnaK-GrpE chaperone cooperation PMID: 25686738
- The nucleotide exchange factor GrpE modulates the chaperone DnaK allosterism. PMID: 25739641
- Dnak and substrate proteins regulate the ATPase activity and dynamics of ClpB. PMID: 25558912
- E. coli DnaK variants with substitutions in subdomains IB and IIB of the nucleotide-binding domain are defective for in vivo and in vitro interactions with ClpB. PMID: 25451597
- Used molecular dynamics simulations, mutagenesis, and enzymatic assays to explore the molecular basis of the structure and function of E coli DnaK. PMID: 24277995
- DnaJ-promoted binding of DnaK to multiple sites on sigma32 in the presence of ATP. PMID: 24532774
- Data indicate that ClpB requires DnaK more stringently than Hsp104 requires Hsp70 for protein disaggregation. PMID: 24280225
- The ATP-bound nucleotide-binding domain of DnaK forms extensive interfaces with a substrate-binding domain that has changed its alpha-helical lid displaced PMID: 23708608
- The DnaK chaperone has evolved to bind peptides in both orientations in the substrate binding cleft with comparable energy without rearrangements of the protein. PMID: 23562829
- Trigger factor dependent refolding of bacterial luciferases in Escherichia coli cells: kinetics, efficiency and effect of the bichaperone system, DnaKJE-ClpB PMID: 23888781
- Disruption of the TF/DnaK chaperone pathway is rescued by overexpression of the redox-regulated chaperone Hsp33. PMID: 23148222
- The "allosterically active" state for the E. coli Hsp70, DnaK, trapped and identified how interactions among the nucleotide-binding domain (NBD), the beta subdomain of the substrate-binding domain (SBD), the SBD alpha-helical lid, and the conserved hydrophobic interdomain linker enable allosteric signal transmission between ligand-binding sites. PMID: 23217711
- The analysis revealed that nearly every protein is predicted to contain multiple DnaK- and DnaJ-binding sites, with the DnaJ sites occurring approximately twice as often. PMID: 22732719
- DnaK oligomers are composed of ordered multimers that are functionally distinct from monomeric DnaK. PMID: 22076723
- The authors demonstrate that DksA expression from a multicopy plasmid is necessary and sufficient for suppression of dnaKJ deletion mutation. PMID: 22267514
- mutation of specific conserved sites within the DnaK C terminus reduces the capacity of the cell to withstand stresses on protein folding caused by elevated temperature or the absence of other chaperones PMID: 21768118
- Results indicate that E. coli Hsp90 and DnaK interact in vivo and in vitro, providing additional evidence to suggest that E. coli Hsp90 and the DnaK system function together PMID: 21525416
- Ribosome assembly defects at very high temperature are partially compensated by plasmid-driven overexpression of the DnaK chaperone. PMID: 21059683
- analysis of complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK) PMID: 20439464
- the Hsp40 J-domain (Jd) shifts DnaK to a client-bound form by stimulating the DnaK ATPase but only when the Jd is brought to DnaK by a client-Hsp40 complex PMID: 20448033
- the interaction of DnaK and its co-chaperones with aggregated substrate is the rate-limiting reaction at the initial steps of disaggregation PMID: 15302880
- results indicate that, despite their high structural identity (approximately 80%) and similar mechanisms of action, the DnaK chaperones of closely related V. harveyi and E.coli bacteria differ functionally. PMID: 15448982
- DnaK-DnaJ-GrpE chaperone system has a role in activating the pi initiator protein PMID: 15485812
- data demonstrates CD154-independent CD40 activation in polymicrobial sepsis and suggests that bacterial HSP70 is capable of stimulating CD40 in vitro and in vivo PMID: 15545825
- Data describe the trigger factor concentration dependence of the glyceraldehyde-3-phosphate dehydrogenase reactivation yield in the presence and absence of the DnaK-DnaJ-GrpE chaperone system in vitro. PMID: 15632130
- Upon oxidative stress, DnaK's N terminus reversibly unfolds in vivo, and DnaK loses its ability to protect proteins against stress-induced aggregation. PMID: 15694339
- DnaK and DnaJ molecular chaperones apparently influence the amount of tryptophanase, the expression of which is regulated at all transcription steps, including transcription elongation. PMID: 15702537
- direct thermal adaptation of the DnaK chaperone system by thermosensing GrpE is essential for efficient chaperone action during heat shock PMID: 15705578
- Results indicate that the complex between DnaK384-638 and its substrate forms a rigid conformation in the beta-domain. PMID: 15752686
- ATPase activity of DnaK is not connected to structural changes of the peptide-binding pocket but rather only has an effect on the LID domain or other further remote residues PMID: 15784262
- chaperones DnaK and GroEL have been identified at the solvent-exposed surface of bacterial inclusion bodies and entrapped within these aggregates, respectively PMID: 15866952
- Separate overproduction of the major chaperone systems, DnaK/DnaJ and GroEL/GroES, established that the former of these is more important in counteracting protein carbonylation. PMID: 15937182
顯示更多
收起更多
-
亞細(xì)胞定位:Cytoplasm. Cell inner membrane; Peripheral membrane protein.
-
蛋白家族:Heat shock protein 70 family
-
數(shù)據(jù)庫鏈接:
KEGG: ecj:JW0013
STRING: 316385.ECDH10B_0014
Most popular with customers
-
Recombinant Human papillomavirus type 16 Protein E7 (E7) (Active)
Express system: E.coli
Species: Human papillomavirus type 16
-
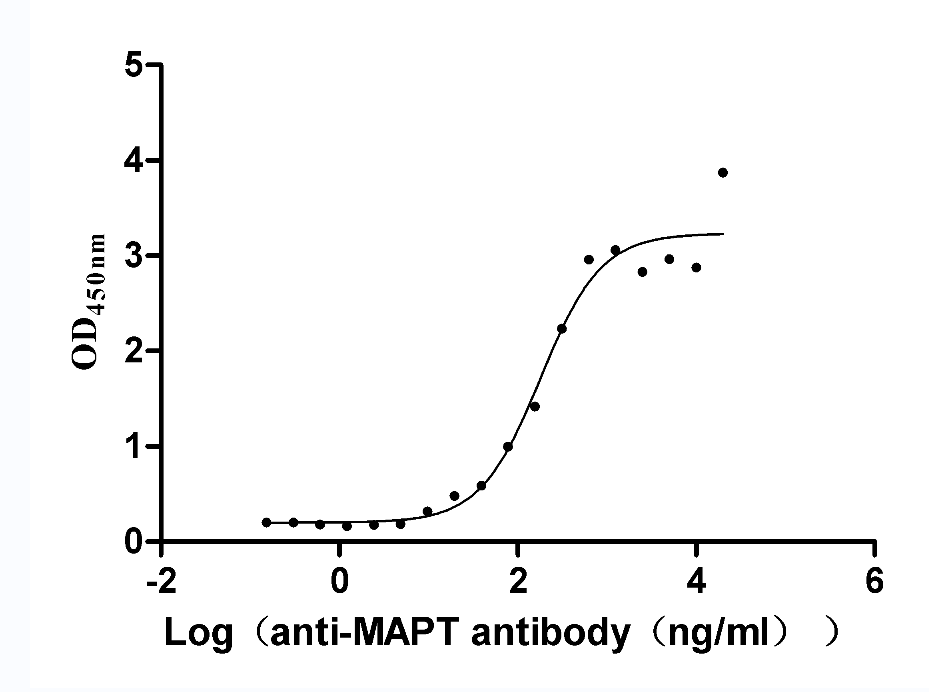
Recombinant Rat Microtubule-associated protein tau (Mapt) (Active)
Express system: Mammalian cell
Species: Rattus norvegicus (Rat)
-
Recombinant Mouse Complement component C1q receptor (Cd93), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
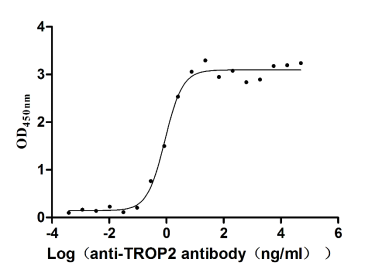
Recombinant Human Tumor-associated calcium signal transducer 2 (TACSTD2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
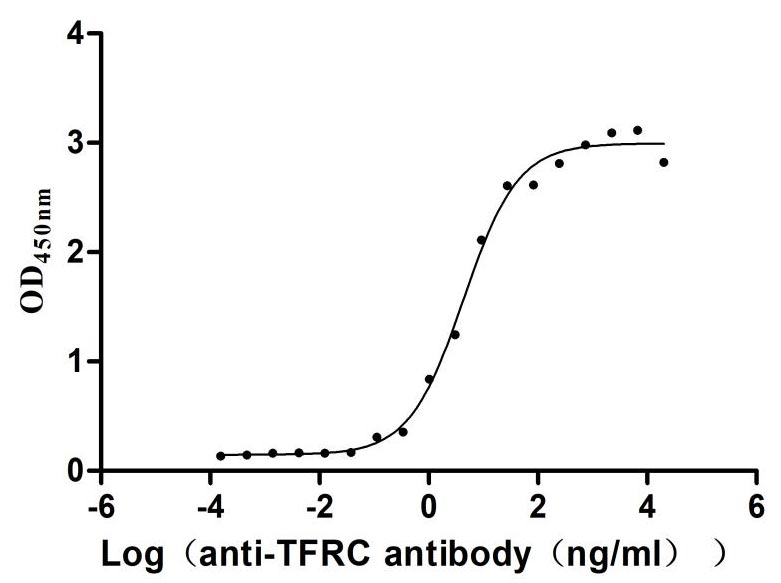
Recombinant Human Transferrin receptor protein 1 (TFRC), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
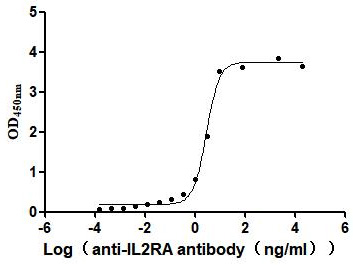
Recombinant Human Interleukin-2 receptor subunit alpha (IL2RA), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
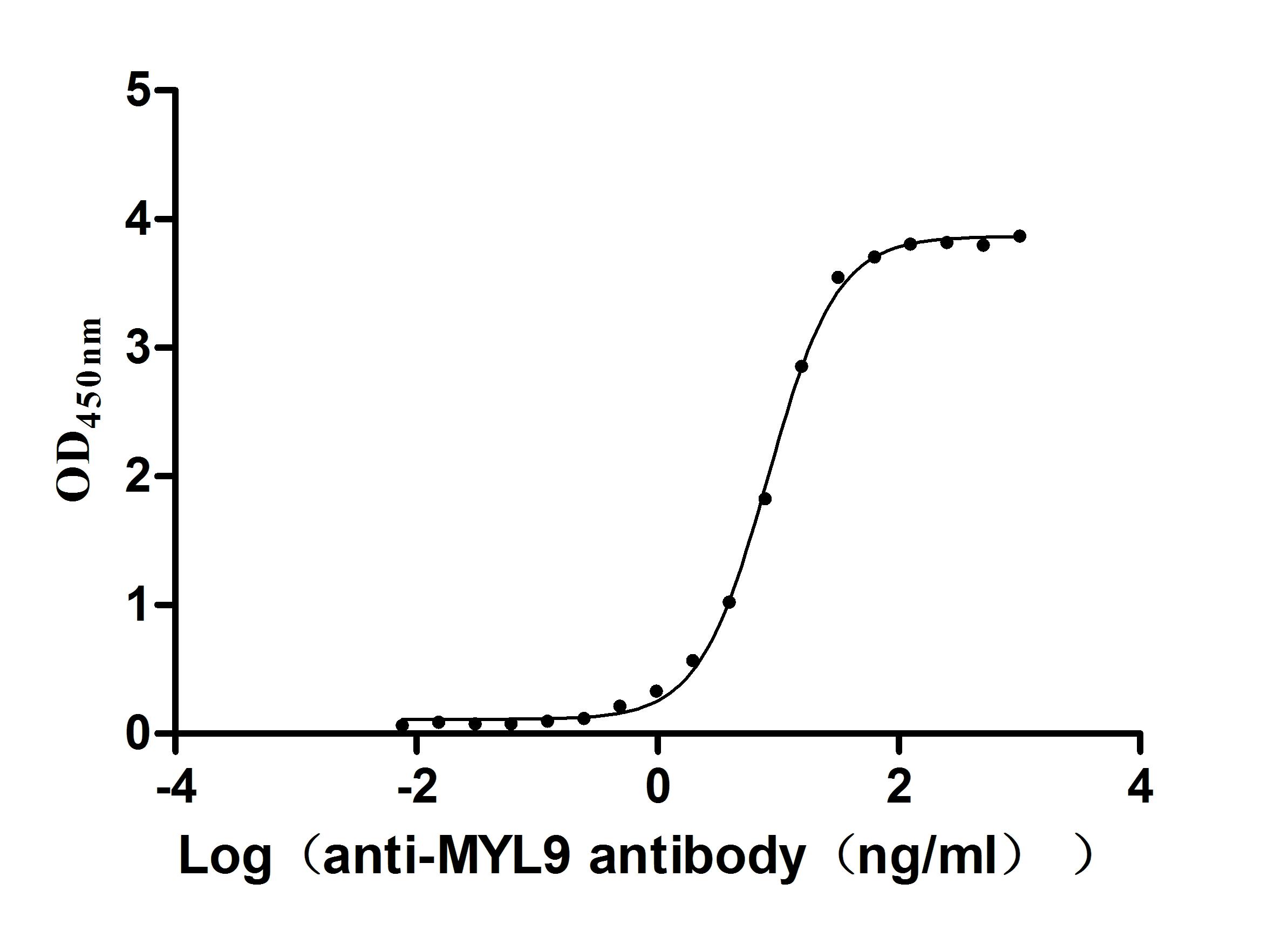
Recombinant Human Myosin regulatory light chain 12B (MYL12B) (Active)
Express system: E.coli
Species: Homo sapiens (Human)
-
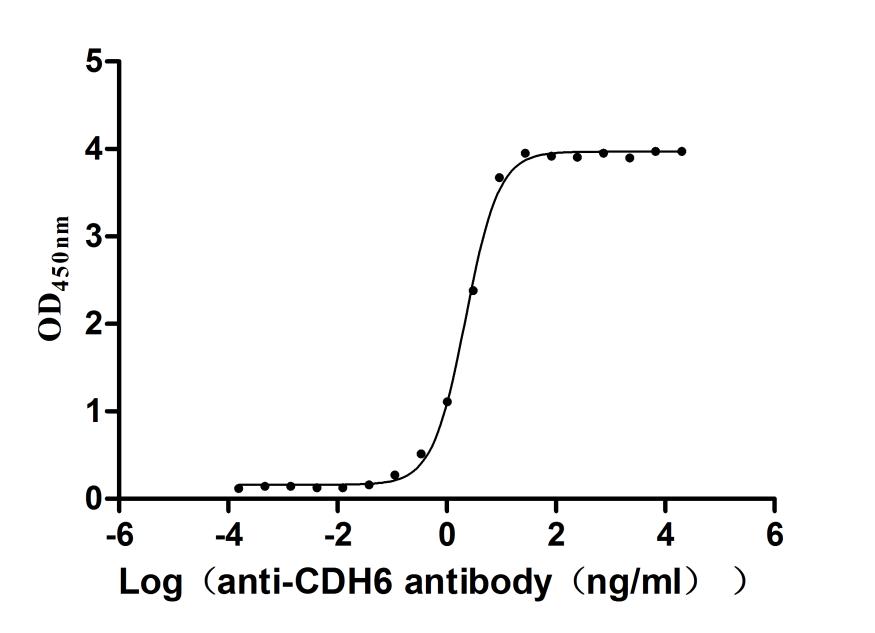
Recombinant Macaca fascicularis Cadherin 6(CDH6),partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)