Recombinant Escherichia coli DNA mismatch repair protein MutS (mutS), partial
-
中文名稱:大腸桿菌mutS重組蛋白
-
貨號:CSB-YP326103ENV
-
規(guī)格:
-
來源:Yeast
-
其他:
-
中文名稱:大腸桿菌mutS重組蛋白
-
貨號:CSB-EP326103ENV
-
規(guī)格:
-
來源:E.coli
-
其他:
-
中文名稱:大腸桿菌mutS重組蛋白
-
貨號:CSB-EP326103ENV-B
-
規(guī)格:
-
來源:E.coli
-
共軛:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名稱:大腸桿菌mutS重組蛋白
-
貨號:CSB-BP326103ENV
-
規(guī)格:
-
來源:Baculovirus
-
其他:
-
中文名稱:大腸桿菌mutS重組蛋白
-
貨號:CSB-MP326103ENV
-
規(guī)格:
-
來源:Mammalian cell
-
其他:
產(chǎn)品詳情
-
純度:>85% (SDS-PAGE)
-
基因名:mutS
-
Uniprot No.:
-
別名:mutS; fdv; b2733; JW2703; DNA mismatch repair protein MutS
-
種屬:Escherichia coli (strain K12)
-
蛋白長度:Partial
-
蛋白標簽:Tag?type?will?be?determined?during?the?manufacturing?process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
產(chǎn)品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
復溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質(zhì)期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事項:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶點詳情
-
功能:This protein is involved in the repair of mismatches in DNA. It is possible that it carries out the mismatch recognition step. This protein has a weak ATPase activity.
-
基因功能參考文獻:
- Mechanism for verification of mismatched and homoduplex DNAs by nucleotides-bound MutS analyzed by molecular dynamics simulations has been reported. PMID: 27238299
- The assembly of an EcMutS-EcMutL-EcMutH search complex illustrates how sequential stable sliding clamps can modulate one-dimensional diffusion mechanics along the DNA to direct mismatch repair PMID: 27851738
- The structure captures MutS in the sliding clamp conformation, where tilting of the MutS subunits across each other pushes DNA into a new channel, and reorientation of the connector domain creates an interface for MutL with both MutS subunits. PMID: 26163658
- MutS and MutL trap recombination intermediates using mismatches formed in the heteroduplex region and secondary DNA structures in the displaced ssDNA, and recruit the UvrD helicase to resolve these trapped intermediates in a directional manner PMID: 23932715
- E. coli MutS has a binding activity specific for non-B form G4 DNA, but such binding appears independent of canonical heteroduplex repair activation. PMID: 22747774
- The results confirm that specifically bound MutS bends DNA at the mismatch. PMID: 22367846
- The Mismatch repair mechanism that involves more MutL accumulation relative to MutS was observed in this study. PMID: 22241777
- analysis of the dynamic MutS-MutL complex formed in DNA mismatch repair in Escherichia coli PMID: 21454657
- mismatch recognition by the MutS proteins is linked to these disparate biological outcomes through regulated interaction of MutL proteins with a wide variety of effector proteins PMID: 20667509
- mutant strains were analyzed using surface plasmon resonance, gel filtration and far-western methods to determine the molecular mechanisms behind the DNA mismatch repair function of MutS PMID: 20953938
- identify a region in the connector domain (domain II) of MutS that binds MutL and is required for mispair-dependent ternary complex formation and mismatch repair PMID: 20080788
- The C-terminal end of MutS is necessary for antirecombination and cisplatin sensitization, but less significant for mutation avoidance during DNA mismatch repair. PMID: 15731339
- MutS glutamate 38 plays a role in both DNA mismatch discrimination and in the authorization of repair. PMID: 16407973
- a beta sliding clamp binds to multiple sites within MutL and MutS PMID: 16546997
- Results support the hypothesis that tetramerization of MutS is important but not essential for MutS function in DNA mismatch repair. PMID: 17012287
- Results show that the interaction of MutY and MutS involves the Fe-S domain of MutY and the ATPase domain of MutS and defend against the mutagenic effect of 8-oxoG lesions in a cooperative manner. PMID: 17114250
- DNA bending could play a role in MutS biochemical modulations induced by a compound lesion and that cisplatin DNA damage signaling by the mismatch repair system could be modulated in a direct mode. PMID: 17400248
- Since movement of MutS along the chain does not require ATP hydrolysis and the ATPase activity is enhanced when MutS dissociates from a heteroduplex, this support model in which ATP binding by MutS induces the formation of a sliding clamp. PMID: 17950245
- The data reveal that the loop plays a direct role in co-ordinating conformational changes involved in long-range communication between Walker box and mismatch recognition domains. PMID: 18673453
- study describes normal-mode calculations of MutS with and without DNA; distinct conformational states are proposed for DNA scanning, mismatch recognition, repair initiation, and sliding along DNA after mismatch recognition PMID: 19254532
- analysis of MutS-, and MutL-dependent activation of MutH and their roles in methyl-directed mismatch repair in vitro PMID: 19783657
顯示更多
收起更多
-
蛋白家族:DNA mismatch repair MutS family
-
數(shù)據(jù)庫鏈接:
KEGG: ecj:JW2703
STRING: 316385.ECDH10B_2901
Most popular with customers
-
Recombinant Human E3 ubiquitin-protein ligase ZNRF3 (ZNRF3), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Lymphocyte antigen 6 complex locus protein G6d (LY6G6D) (Active)
Express system: Yeast
Species: Homo sapiens (Human)
-
Recombinant Human Early activation antigen CD69 (CD69), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Myosin regulatory light polypeptide 9 (MYL9) (Active)
Express system: Yeast
Species: Homo sapiens (Human)
-
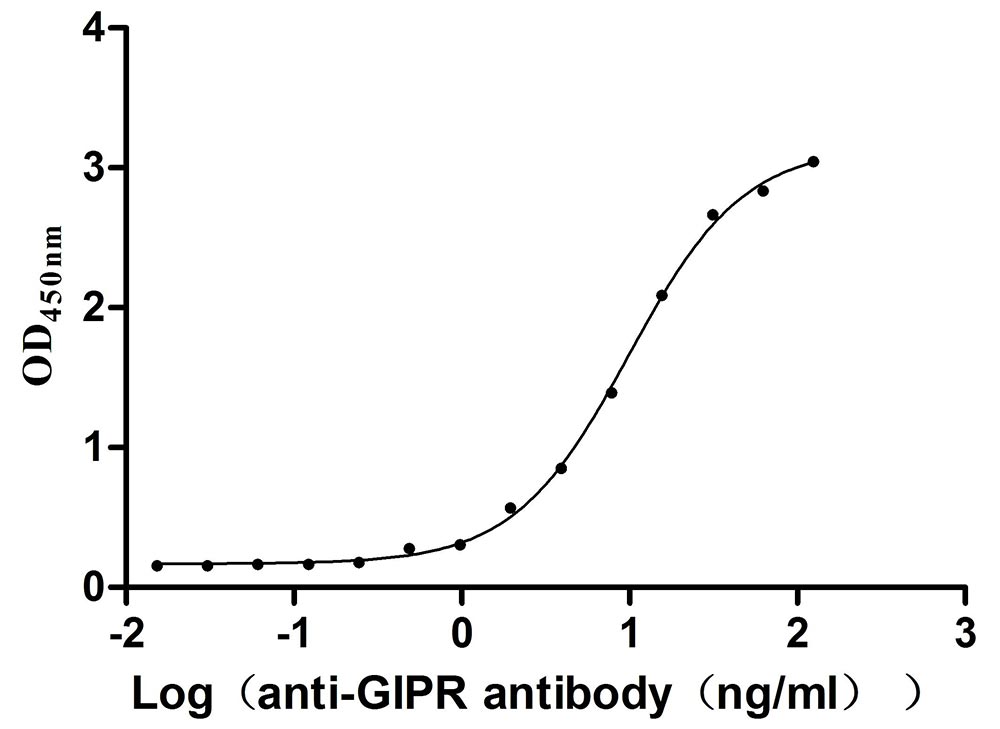
Recombinant Mouse Gastric inhibitory polypeptide receptor (Gipr), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
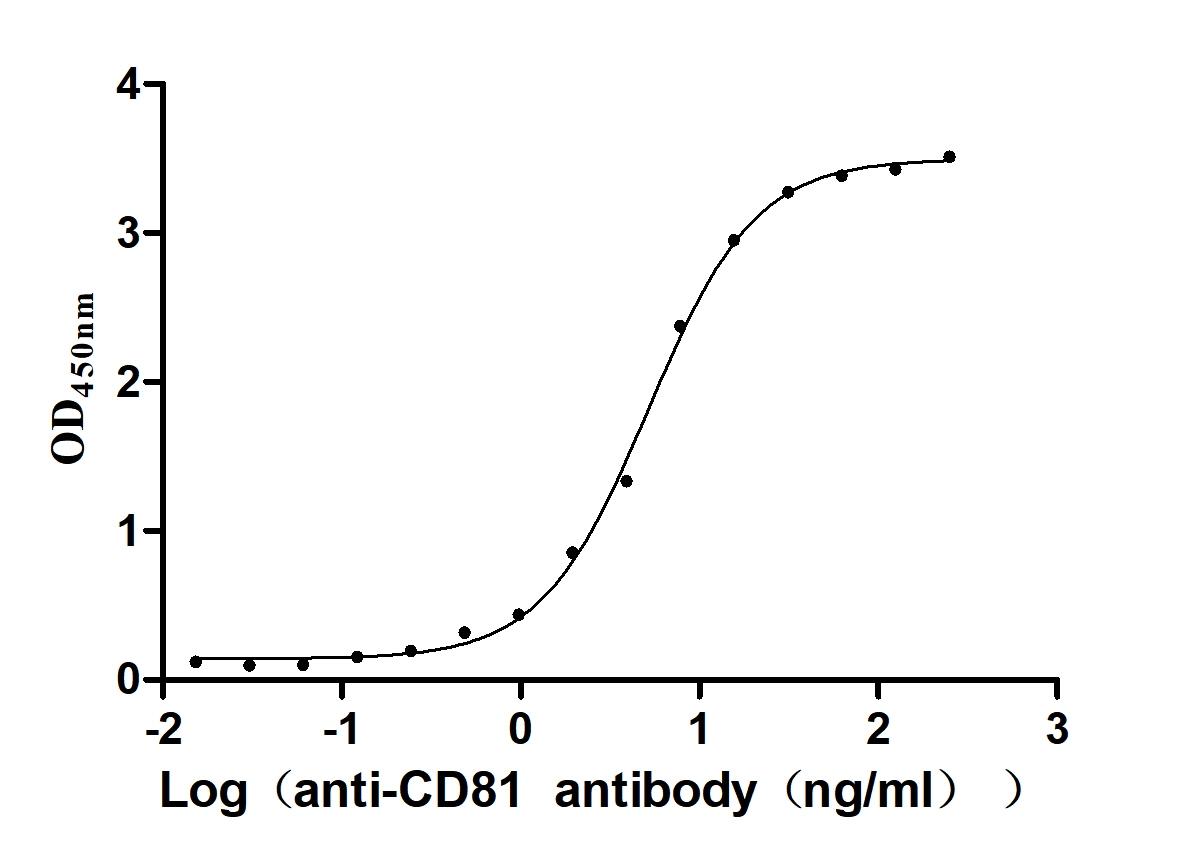
Recombinant Human CD81 antigen (CD81), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
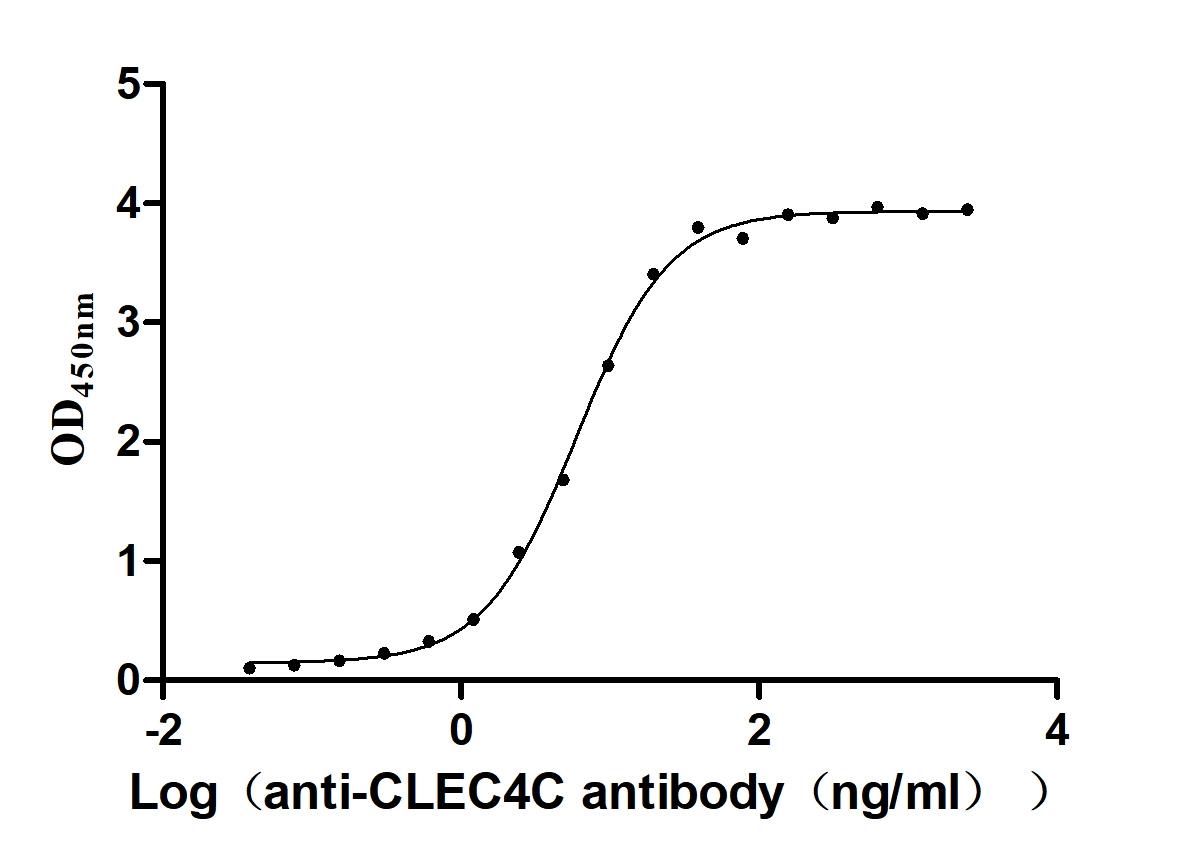
Recombinant Macaca fascicularis C-type lectin domain family 4 member C(CLEC4C), partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
Recombinant Human C-C chemokine receptor type 6(CCR6)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)