Recombinant Saccharomyces cerevisiae DNA-directed RNA polymerase II subunit RPB1 (RPO21), partial
-
中文名稱:釀酒酵母RPO21重組蛋白
-
貨號:CSB-YP018327SVG
-
規(guī)格:
-
來源:Yeast
-
其他:
-
中文名稱:釀酒酵母RPO21重組蛋白
-
貨號:CSB-EP018327SVG
-
規(guī)格:
-
來源:E.coli
-
其他:
-
中文名稱:釀酒酵母RPO21重組蛋白
-
貨號:CSB-EP018327SVG-B
-
規(guī)格:
-
來源:E.coli
-
共軛:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名稱:釀酒酵母RPO21重組蛋白
-
貨號:CSB-BP018327SVG
-
規(guī)格:
-
來源:Baculovirus
-
其他:
-
中文名稱:釀酒酵母RPO21重組蛋白
-
貨號:CSB-MP018327SVG
-
規(guī)格:
-
來源:Mammalian cell
-
其他:
產(chǎn)品詳情
-
純度:>85% (SDS-PAGE)
-
基因名:RPO21
-
Uniprot No.:
-
別名:RPO21; RPB1; RPB220; SUA8; YDL140C; D2150; DNA-directed RNA polymerase II subunit RPB1; RNA polymerase II subunit 1; RNA polymerase II subunit B1; EC 2.7.7.6; DNA-directed RNA polymerase III largest subunit; RNA polymerase II subunit B220
-
種屬:Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
-
蛋白長度:Partial
-
蛋白標簽:Tag?type?will?be?determined?during?the?manufacturing?process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
產(chǎn)品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
復溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質(zhì)期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事項:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶點詳情
-
功能:DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Largest and catalytic component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Forms the polymerase active center together with the second largest subunit. Pol II is the central component of the basal RNA polymerase II transcription machinery. During a transcription cycle, Pol II, general transcription factors and the Mediator complex assemble as the preinitiation complex (PIC) at the promoter. 11-15 base pairs of DNA surrounding the transcription start site are melted and the single-stranded DNA template strand of the promoter is positioned deeply within the central active site cleft of Pol II to form the open complex. After synthesis of about 30 bases of RNA, Pol II releases its contacts with the core promoter and the rest of the transcription machinery (promoter clearance) and enters the stage of transcription elongation in which it moves on the template as the transcript elongates. Pol II appears to oscillate between inactive and active conformations at each step of nucleotide addition. Elongation is influenced by the phosphorylation status of the C-terminal domain (CTD) of Pol II largest subunit (RPB1), which serves as a platform for assembly of factors that regulate transcription initiation, elongation, termination and mRNA processing. Pol II is composed of mobile elements that move relative to each other. The core element with the central large cleft comprises RPB3, RBP10, RPB11, RPB12 and regions of RPB1 and RPB2 forming the active center. The clamp element (portions of RPB1, RPB2 and RPB3) is connected to the core through a set of flexible switches and moves to open and close the cleft. A bridging helix emanates from RPB1 and crosses the cleft near the catalytic site and is thought to promote translocation of Pol II by acting as a ratchet that moves the RNA-DNA hybrid through the active site by switching from straight to bent conformations at each step of nucleotide addition. In elongating Pol II, the lid loop (RPB1) appears to act as a wedge to drive apart the DNA and RNA strands at the upstream end of the transcription bubble and guide the RNA strand toward the RNA exit groove located near the base of the largely unstructured CTD domain of RPB1. The rudder loop (RPB1) interacts with single-stranded DNA after separation from the RNA strand, likely preventing reassociation with the exiting RNA. The cleft is surrounded by jaws: an upper jaw formed by portions of RBP1, RPB2 and RPB9, and a lower jaw, formed by RPB5 and portions of RBP1. The jaws are thought to grab the incoming DNA template, mainly by RPB5 direct contacts to DNA.
-
基因功能參考文獻:
- Here, the authors identify a site of ubiquitination (K1246) in the catalytic subunit of RNAPII close to the DNA entry path. Ubiquitination was increased in the absence of the Bre5-Ubp3 ubiquitin protease complex. PMID: 29027900
- The authors determined that the tandem SH2 domain of Saccharomyces cerevisiae Spt6 binds the linker region of the RNA polymerase II subunit Rpb1 rather than the expected sites in its heptad repeat domain. The 4 nM binding affinity requires phosphorylation at Rpb1 S1493 and either T1471 or Y1473. PMID: 28826505
- Data suggest the binding motif in the RPB1 (RPO21) switch 1 of RNA polymerase II (pol II) can sense the structural changes of the DNA minor groove (minor groove sensor) before duplex unwinding. PMID: 27791148
- RPO21 interaction with LYS2 and Spt elongation factors PMID: 27261007
- Promoter and template-specific effects on severity of gene expression defects have been identified for both fast and slow Pol II mutants. PMID: 28119420
- Studies indicate that the covalent, flexible linker ensured that transcription factor TFIIB (TFIIB) was recruited stoichiometrically near its binding site on the RNA polymerase (Pol) II (Pol II) surface. PMID: 27528766
- RPB1 mutants in the foot region of RNA polymerase II affect the assembly of the complex by altering the correct association of both the Rpb6 and the Rpb4/7 dimer PMID: 27001033
- Sen1, Nrd1 and Ysh1 play a role in RNA polymerase II termination. PMID: 25299594
- Results indicate that multiple mechanistic features contribute to 5'-3' exoribonuclease Rat1-mediated termination of RNA polymerase II (RNAPII). PMID: 25722373
- The results show that the Rpb4/7 heterodimer in Saccharomyces cerevisiae plays a key role in controlling phosphorylation of the carboxy terminal domain of the Rpb1 subunit of RNA polymerase II. PMID: 25416796
- Data indicate reduced promoter association of Mediator and TATA-binding protein in a RNA polymerase II (rpb1-1) mutant. PMID: 24727477
- These results show that, with some spatial constraints, CTD can function even when disconnected from Rpb1. PMID: 24035501
- Results indicate an important role of the intrinsic blocks to forward translocation in pausing by RNA polymerase II (RNAP II). PMID: 23238253
- RPB1 (RPO21) mutations increase transcriptional slippage in S. cerevisiae PMID: 23223234
- Data show that polymerases transcribing opposite DNA strands cannot bypass each other, and head-to-head collision results in RNA polymerase II (RNAPII) stopping. PMID: 23041286
- find that the transcription processivity defect imposed on RNAPII by the rpb1-N488D mutation is corrected upon Rat1p inactivation. PMID: 20188675
- RPB1 has a role in hypersensitivity to 6-azauracil PMID: 16510790
- An Rsp5 dimer assembled on the RNA polymerase II (RNAP II) C-terminal domain; ubiquitylation sites bind Ubc5, which in turn binds Rsp5 to allow modification of RPB1 subunit of RNAP II. PMID: 17418786
- In response to ultraviolet radiation, Rpb9 also functions in promoting ubiquitylation and degradation of Rpb1, the largest subunit of Pol II. PMID: 17452455
- Saccharomyces cerevisiae RNA polymerase II (Pol II) "trigger loop" participates in substrate selection PMID: 18538653
- mutant showed a 10-fold decrease in fidelity of transcription elongation in vitro PMID: 18538654
- Rpb1p interacts with a large number of proteins involved in mRNA synthesis, processing, export, and other cellular processes. PMID: 19193966
- results demonstrate a novel covalent modification of Rpb1 in response to UV induced DNA damage or transcriptional impairment, and unravel an important link between the modification and the DNA damage checkpoint response PMID: 19384408
- Data show that Rpb1 CTD serine 7 is also phosphorylated and that Ser-7(P) chromatin immunoprecipitation patterns resemble those of Ser-5(P). PMID: 19679665
顯示更多
收起更多
-
亞細胞定位:Nucleus.
-
蛋白家族:RNA polymerase beta' chain family
-
數(shù)據(jù)庫鏈接:
KEGG: sce:YDL140C
STRING: 4932.YDL140C
Most popular with customers
-
Recombinant Human Prolactin receptor (PRLR), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Angiopoietin-2 (ANGPT2) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Microtubule-associated protein tau (MAPT) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
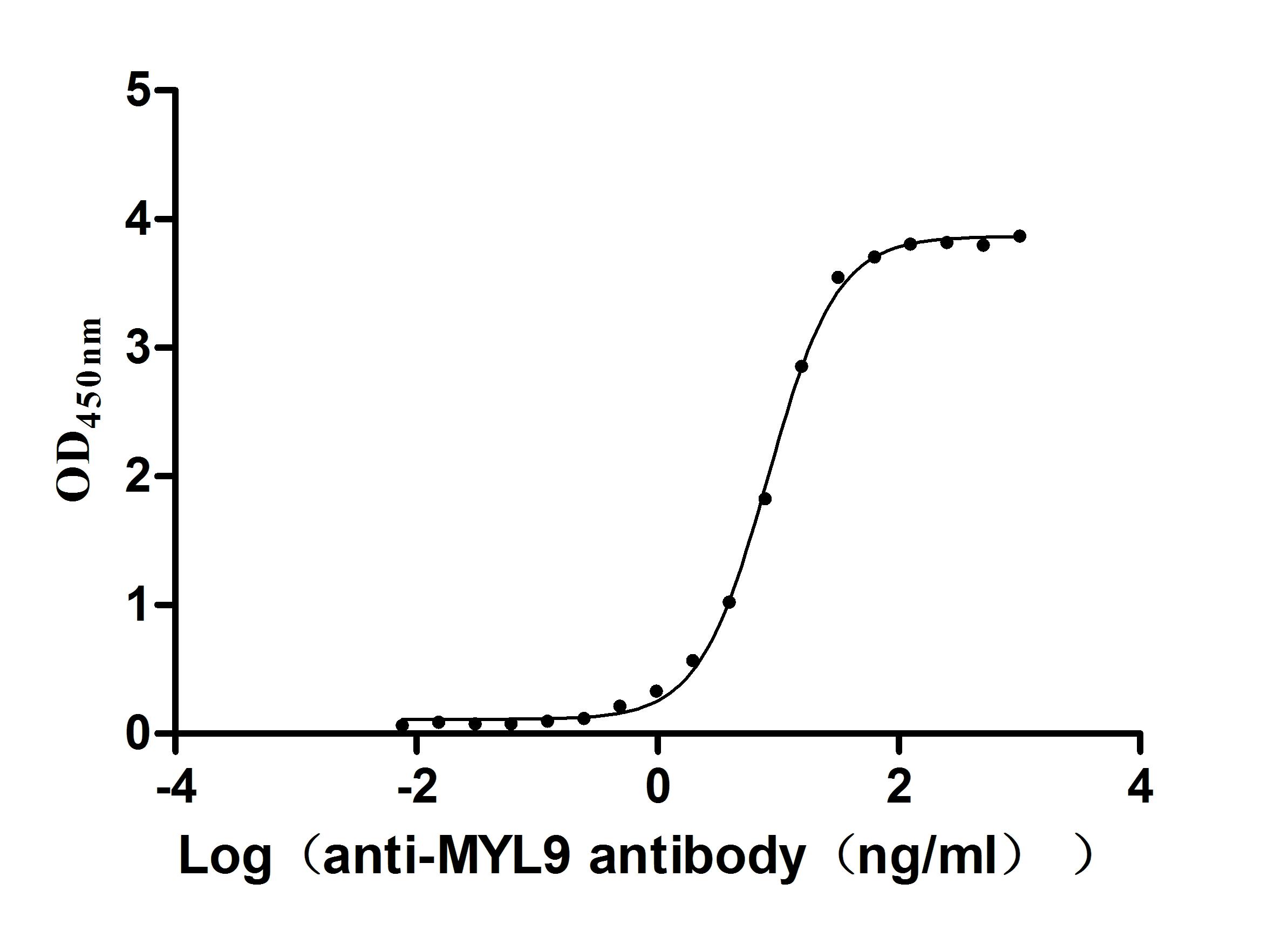
Recombinant Human Myosin regulatory light chain 12B (MYL12B) (Active)
Express system: E.coli
Species: Homo sapiens (Human)
-
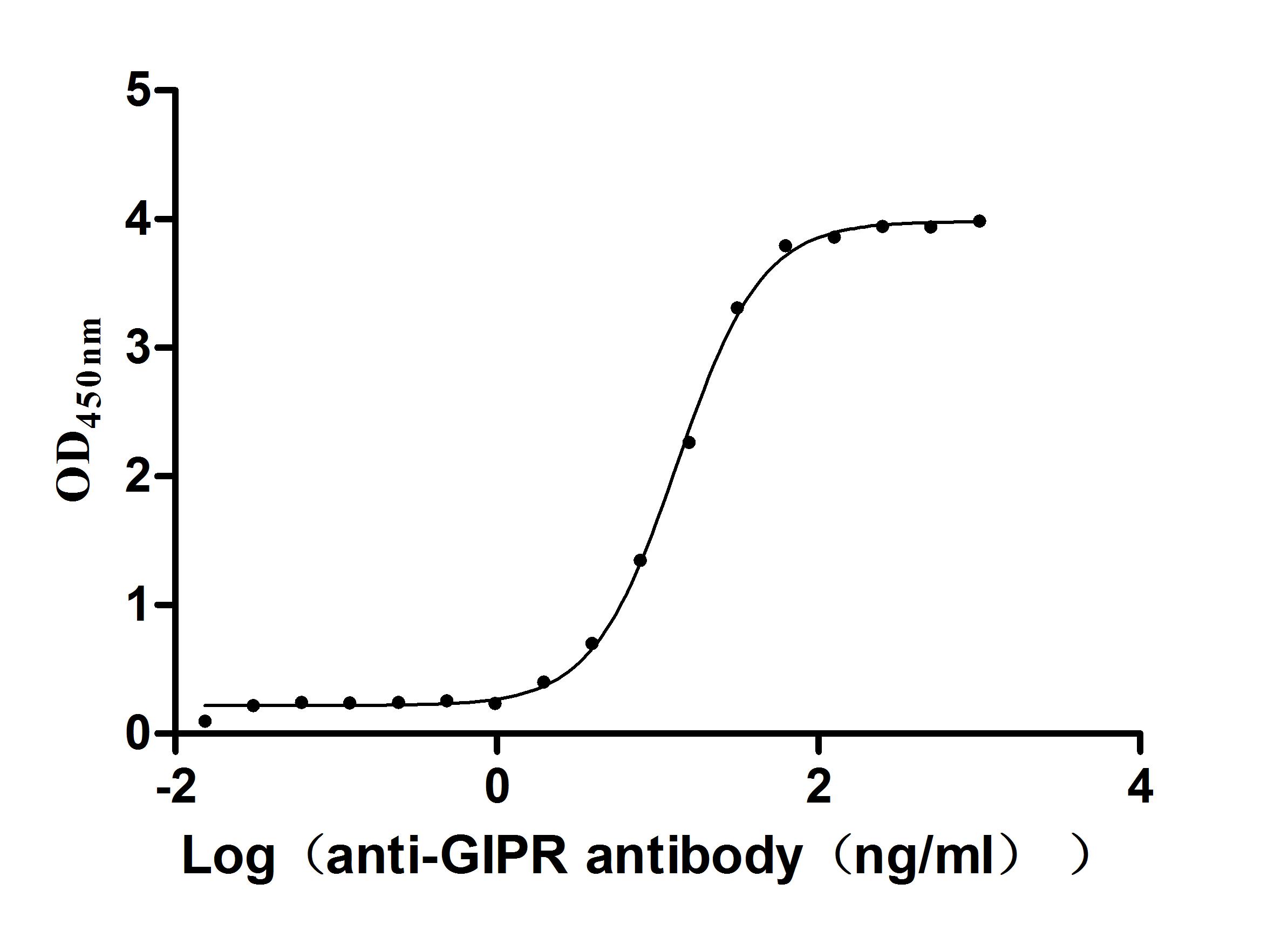
Recombinant Macaca fascicularis Gastric inhibitory polypeptide receptor (GIPR), partial (Active)
Express system: yeast
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
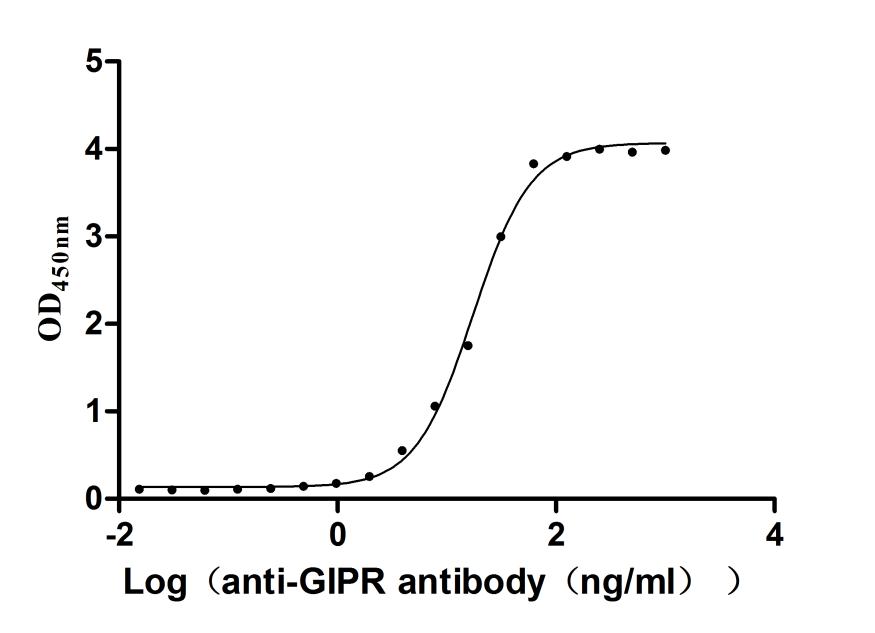
Recombinant Human Gastric inhibitory polypeptide receptor(GIPR),partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Urokinase-type plasminogen activator(PLAU) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)


-AC1.jpg)
-AC1.jpg)